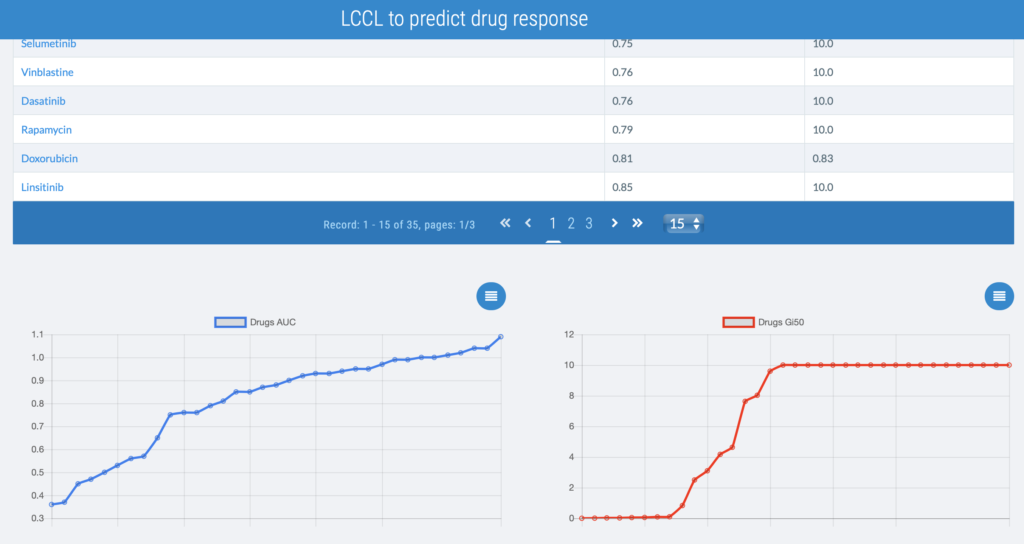
New publication in Nature Rev. Gastroenterol. Hepatol. by JC Nault, Massih Ningarhari, et al

ABSTRACT
Telomerase is a key enzyme for cell survival that prevents telomere shortening and the subsequent cellular senescence that is observed after many rounds of cell division. In contrast, inactivation of telomerase is observed in most cells of the adult liver. Absence of telomerase activity and shortening of telomeres has been implicated in hepatocyte senescence and the development of cirrhosis, a chronic liver disease that can lead to hepatocellular carcinoma (HCC) development. During hepatocarcinogenesis, telomerase reactivation is required to enable the uncontrolled cell proliferation that leads to malignant transformation and HCC development. Part of the telomerase complex, telomerase reverse transcriptase, is encoded by TERT, and several mechanisms of telomerase reactivation have been described in HCC that include somatic TERT promoter mutations, TERTamplification, TERT translocation and viral insertion into the TERTgene. An understanding of the role of telomeres and telomerase in HCC development is important to develop future targeted therapies and improve survival of this disease. In this Review, the roles of telomeres and telomerase in liver carcinogenesis are discussed, in addition to their potential translation to clinical practice as biomarkers and therapeutic targets.