Functional Genomics of Liver Tumors
FunGeST lab – an insight into liver tumors genomics
Group Leader
Actual Projects
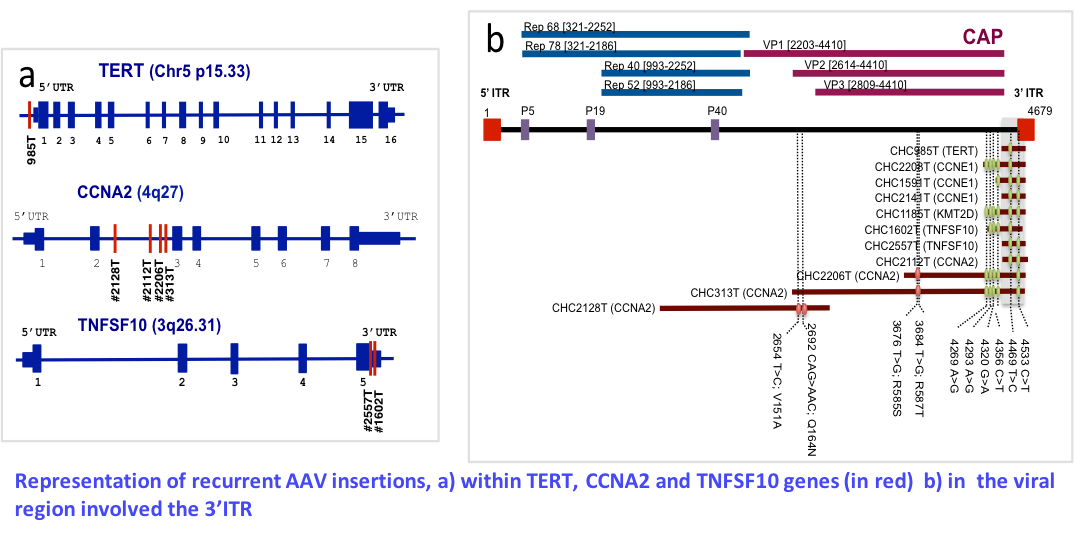
Role of viral infection and integration in liver carcinogenesis
Collaborators: S Imbeaud (IR), C Peneau (PhD st), T La Bella (PhD st), J Zucman-Rossi (PUPH);
Involvement of pathogen agents in human carcinogenesis remain to be investigated. Our recent identification of AAV2 as the fourth virus involved in insertional mutagenesis prompt us to develop a project to evaluate consequences of HBV, HCV and AAV infections in liver tumorigenesis by integrating the analysis of viral and tumor genome alterations. Our aim is to reconstruct the natural history of the viruses (genome variation, replication, structural alterations, selection…) in the context of tumor cell selection in patients putatively treated by antiviral therapies or exposed to multiple pathogens. We will also search for additional pathogens in WGS data and collaborate with the international ICGC viruses project.
Fundings: LNCC, ANRS, Biomerieux Foundation
New molecular subtypes and cancer driver genes in pediatric liver tumors
Collaborators: G Morcrette (PhD st), E Letouzé (CR Inserm), J Pilet (PhD st), S Imbeaud (IR), G Couchy (IE, PhD st), Julien Calderaro (pathologist), JC Nault (MCUPH), J Zucman-Rossi (PUPH); in cooperation with SIOPEL,
Most of pediatric liver neoplasms are poorly analyszed at the genomic and molecular level. They include hepatocellular carcinomas (HCC), fibrolamellar carcinomas (FLC), hepatoblastomas (HB), transitional liver cell tumors (TLCT) and also benign neoplasms, hepatocellular adenomas (HCA) and focal nodular hyperplasias (FNH). We aim to perform an integrated genomic analysis of these pediatric hepatocellular tumors to better understand their mechanism of tumorigenesis. To this purpose, we have collected a series of 193 patients with pediatric frozen liver tumors part of the national HEPATOBIO tumor resource. Within this series, 48 cases/67 frozen tumor samples have been selected for whole genome sequencing (WGS) and RNA sequencing (RNAseq) within the GEPELIN project funded by France Génomique for the WGS part. Our specific aims are to (1) identify putative driver genes altered in pediatric tumors and validate them functionally (2) perform new transcriptomic classification to derive new altered pathways and diagnostic/prognostic markers, (3) characterize the mutational and chromosome rearrangement signatures to identify new mechanism of carcinogenesis (4) search for therapeutic targets. These results will be compared with the adult liver tumors data that we have accumulated in the lab.
Fundings: LNCC, France Genomique.
Innovative genomic data integration to decipher the interaction between risk factors, endogeneous cellular processes and genomic alterations in liver carcinogenesis
Collaborators: E Letouzé , J Shinde , S Imbeaud (IR), L Meunier (PhD st), Q Bayard (M2), T Hirsch (Post-doc), J Zucman-Rossi (PUPH
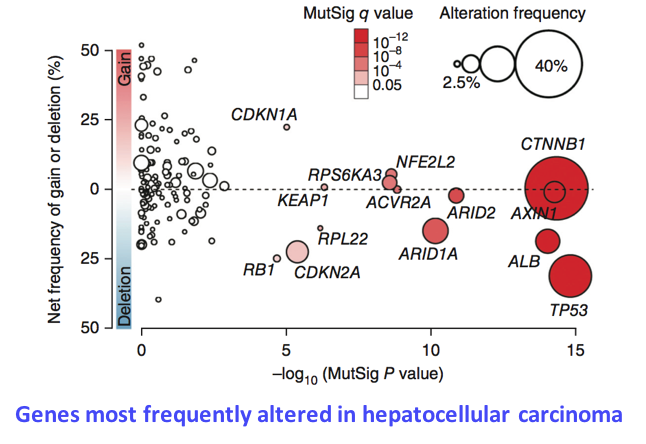
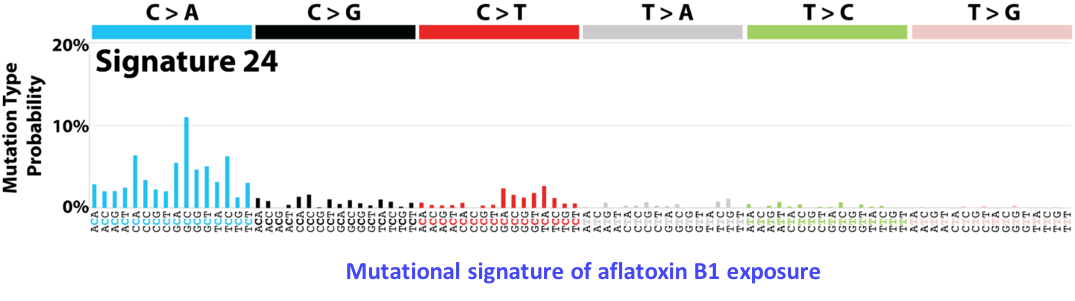
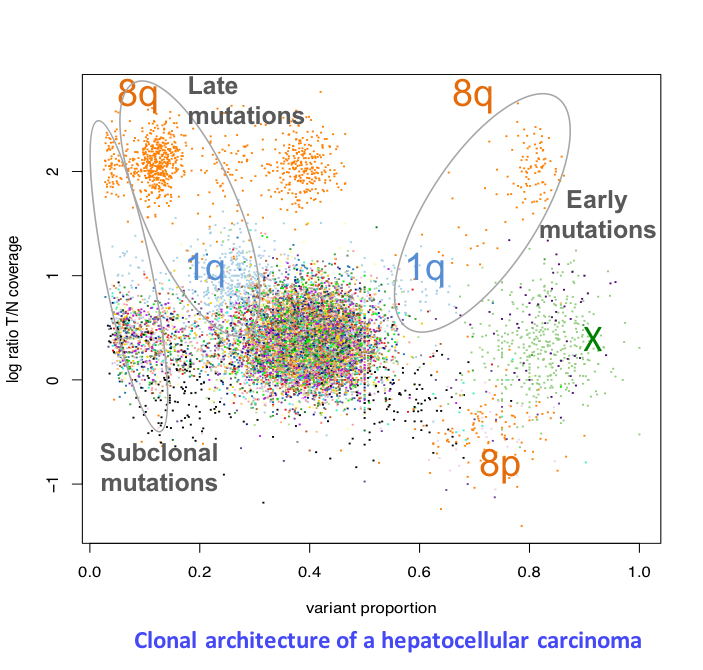
In the last 5 years, we have generated large genomic data sets including 350 tumors analyzed by whole exome sequencing, 270 by RNA-seq, 250 by methylation arrays and 100 by whole genome sequencing. We will use this exceptional resource to unravel key questions in the natural history of liver cancers. (1) What mutational processes drive tumorigenesis? We have so far identified 10 point-mutation signatures associated with known (aflatoxin B1, tobacco) or unknown mutagenic processes in liver cancers. We will extend these analyses to signatures of indels and structural rearrangements, and perform a meta-analysis of all published liver cancer data sets to unravel the association of signatures with risk factors and predisposing variants. (2) What are the missing drivers in liver cancers? Analysis of coding mutations by us and others revealed tens of driver genes and pathways but 30% of tumors still have no identified driver event. In order to identify non-coding driver alterations, we will use whole genome and RNA-seq data to systematicallly screen for mutations and structural rearrangements modifying regulatory regions, chromatin context and 3’ UTR sequences and affecting mRNA expression and stability. (3) How to the genomic, epigenomic and transcriptional layers interact in tumor cells? We will develop innovative strategies to unravel the connexions between genomic alterations, DNA methylation and gene expression profiles. (4) What is the timing of genomic alterations along tumorigenesis? We and others have developed statistical approaches to time mutational signatures, copy-number alterations and driver events in the life history of a cancer using intra-tumor heterogeneity. We will conduct an ambitious project to analyze the evolution of genomic, transcriptional and micro-environment features in 25 patients with multiple samples along the treatment.
Fundings: Cancer environment, HTE HETCOLI, France Genomique, Canceropole
Genetic predisposition to benign and malignant liver tumors
E Letouzé (CR Inserm), P Nahon (PUPH), E Trepo (Post-doc), J Yang (Ph st), JC Nault (MCUPH), Zucman-Rossi (PUPH)
Candidate gene studies have uncovered a limited number of variants reproducibly linked to hepatocellular carcinoma (HCC). The HECAM project aims to identify newgermline variants predisposing to HCC in the French population and to test their ability to modify patient care in clinical practice. Therefore, a genome-wide association study will be performed in a multicenter cohort of chronic liver disease patients with (n = 2,066) and without (n = 2,666) HCC using the Illumina Infinium Global Screening Array including ~ 660,000 markers). Validation of the top variants will be performed in two French replication cohorts of cirrhotic patients included in HCC surveillance programs (n = 2,249). The performance for HCC risk stratification will be finally assessed through integration of genetic information into specific algorithm-based prediction models. An original integrative approach including somatic information already generated will be used for prioritizing variants impacting liver carcinogenesis. Finally, genotype-phenotype associations will be performed to build prognostic models. We also plan to analyze benign liver tumors to search for genetic predisposition to the development of hepatocellular adenoma in a cohort of 500 patients with in parallel an epidemiological study.
Fundings: HECAM, ANRS, AFEF
Team
Biologists
Clinicians
Fundings
![]()
Latest Publications
APC germline hepatoblastomas demonstrate cisplatin-induced intratumor tertiary lymphoid structures. Morcrette G, Hirsch TZ, Badour E, Pilet J, Caruso S, Calderaro J, Martin Y, Imbeaud S, Letouzé E, Rebouissou S, Branchereau S, Taque S, Chardot C, Guettier C, Scoazec JY, Fabre M, Brugières L, Zucman-Rossi J. Oncoimmunology. 2019 Mar 28;8(6):e1583547. doi: 10.1080/2162402X.2019.1583547. eCollection 2019.
Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Dhanasekaran R, Nault JC, Roberts LR, Zucman-Rossi J. Gastroenterology. 2019 Jan;156(2):492-509. doi: 10.1053/j.gastro.2018.11.001. Epub 2018 Nov 4. Review.
Systemic AA Amyloidosis Caused by Inflammatory Hepatocellular Adenoma. Calderaro J, Letouzé E, Bayard Q, Boulai A, Renault V, Deleuze JF, Bestard O, Franco D, Zafrani ES, Nault JC, Moutschen M, Zucman-Rossi J. N Engl J Med. 2018 Sep 20;379(12):1178-1180. doi: 10.1056/NEJMc1805673.
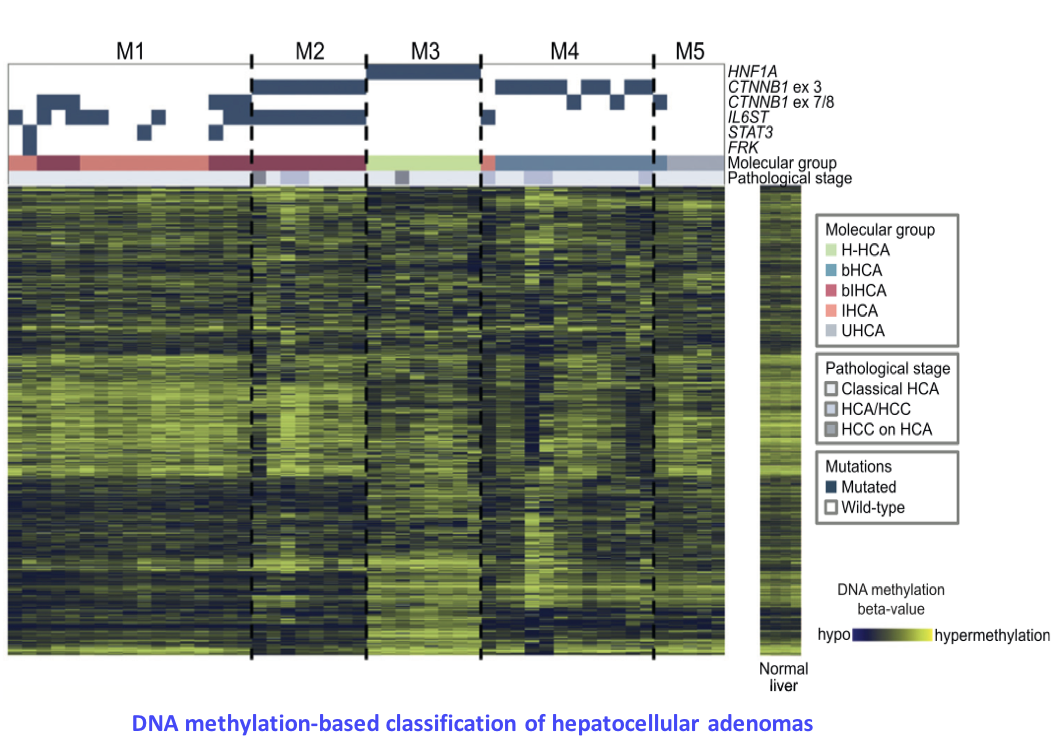
Argininosuccinate synthase 1 and periportal gene expression in sonic hedgehog hepatocellular adenomas. Nault JC, Couchy G, Caruso S, Meunier L, Caruana L, Letouzé E, Rebouissou S, Paradis V, Calderaro J, Zucman-Rossi J. Hepatology. 2018 Sep;68(3):964-976. doi: 10.1002/hep.29884. Epub 2018 Jun 6
Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Letouzé E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J, Bioulac-Sage P, Prévôt S, Azoulay D, Paradis V, Imbeaud S, Deleuze JF, Zucman-Rossi J. Nat Commun. 2017 Nov 3;8(1):1315
Pro-angiogenic gene expression is associated with better outcome on sunitinib in metastatic clear-cell renal cell carcinoma. Beuselinck B, Verbiest A, Couchy G, Job S, de Reynies A, Meiller C, Albersen M, Verkarre V, Lerut E, Méjean A, Patard JJ, Laguerre B, Rioux-Leclercq N, Schöffski P, Oudard S, Zucman-Rossi J. Acta Oncol. 2018 Apr;57(4):498-508. doi: 10.1080/0284186X.2017.1388927. Epub 2017 Nov 2