Our Public Data
2019
Gastroenterology – Caruso et al – Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response.
| Item | Data | Details |
|---|---|---|
| Cell lines | LCCL_description | Description of liver cancer cell lines. |
| Cell lines | Somatic_alterations | List of putative somatic genomic alterations identified in 34 Liver Cancer Cell Lines by whole-exome sequencing. |
| Cell lines | Fusion_transcripts | Fusion transcripts identified in 34 liver cancer cell lines by RNA-sequencing. |
| Cell lines | HBV_fusions | HBV RNA fusions identified by RNA-sequencing among 34 liver cancer cell lines. |
| Cell lines | Drivers | List of the 252 cancer driver genes used to analyze association between drug sensitivity and genetic alterations in 34 liver cancer cell lines. |
| Cell lines | RNAseq_vsn | RNA expression in 34 liver cancer cell lines (vsn) |
| Cell lines | RNAseq_FPMK | RNA expression in 34 liver cancer cell lines (FPKM) |
| Cell lines | microRNAseq_vsn | microRNA expression in 34 liver cancer cell lines (vsn) |
| Cell lines | microRNAseq_RPM | microRNA expression in 34 liver cancer cell lines (RPM) |
| Cell lines | Protein_expression | Expression of 126 proteins analyzed by RPPA in 34 liver cancer cell lines (log2-transformed). |
| Cell lines/Drugs | AUC | Drug sensitivity in 34 liver cancer cell lines. Table of AUC values for each drug |
| Cell lines/Drugs | GI50 | Drug sensitivity in 34 liver cancer cell lines. Table of GI50 values for each drug |
| Drugs | Screened_compounds | Description of screened compounds. |
Read More
Molecular & Cellular Analysis Instruments
Advanced Technologies within FunGeST lab
Persons in charge
 Illumina MiSeq
Illumina MiSeq
Manufacturer’s description: The MiSeq desktop sequencer allows you to access more focused applications such as targeted gene sequencing, metagenomics, small genome sequencing, targeted gene expression, amplicon sequencing, and HLA typing. New MiSeq reagents enable up to 15 Gb of output with 25 M sequencing reads and 2×300 bp read lengths.
Applications currently used: Targeted sequencing (paired-end 2x150bp) using multiplexed PCR for library preparation.

Applied Biosystems 3500 XL Genetic Analyzer (24-Capillary arrays)
Manufacturer’s description: Based on one of the most widely used, widely trusted sequencing methodologies available (Sanger sequencing) the 3500 Series Genetic Analyzer is designed to deliver the accuracy you demand. The 3500 platform can run a wide variety of applications, including de novo sequencing and resequencing (mutational profiling), microsatellite analysis, MLPA, AFLP, LOH, MLST, and SNP validation or screening.
Applications currently used: Sanger sequencing (read length up to 1000bp)

Nanostring nCounter Analysis System
Manufacturer’s description: The nCounter® Analysis System offers a simple, cost-effective way to profile hundreds of mRNAs, microRNAs, or DNA targets simultaneously with high sensitivity and precision. The digital quantification of target molecules and high levels of multiplexing eliminate the compromise between data quality and data quantity, producing excellent sensitivity and high reproducibility for studies of hundreds of targets. The system uses molecular “barcodes” and single molecule imaging to quantitate up to 800 unique transcripts in a single reaction.
Applications currently used: Gene expression and miRNA analysis on FFPE tumor samples
Fluidigm Biomark HD System
Controller AX (Access Array 48.48), Controller HX (Dynamic Array 96.96), Controller MX (Dynamic Array 48.48)
Manufacturer’s description: Fluidigm’s revolutionary integrated fluidic circuits (IFCs) empower life science research by automating PCR reactions in nanoliter volumes. This means using less sample and reagent, and a single microfluidic device, to achieve the high-quality, consistent results your work depends on. The Biomark HD system runs IFCs in either real-time or end-point read modes, bringing flexible, efficient and economical PCR solutions to a range of applications such as digital PCR, gene expression, genotyping, library preparation for next generation sequencing.
Application currently used: Gene expression, genotyping and library preparation
Applied Biosystems 7900 HT Fast Real-Time PCR System
Manufacturer’s description: The Applied Biosystems 7900HT Fast Real-Time PCR System is the only real-time quantitative PCR system that combines 384-well plate compatibility with fully automated robotic loading. Acknowledged as the gold standard in real-time PCR, the 7900HT system combined with TaqMan®Assays enables you to achieve unprecedented throughput and flexibility, allowing you to pursue projects beyond the scope of previous real-time instruments
Application currently used: Gene expression analysis, allelic discrimination, library quantification

Operetta CLS High-Content Analysis System
Manufacturer’s description: The Operetta CLS system combines speed and sensitivity with the powerful and intuitive data analysis. It is a combination of technologies with a powerful, stable 8x LED light source for optimal excitation of fluorophores and confidence in results. It contains also a proprietary automated water-immersion objectives with very high numerical aperture enable high resolution and fast read times with minimal photodamage. The confocal spinning disk technology provides a fast and gentle imaging process, enabling efficient background rejection, live cell experiments, and 3D imaging. Its large format sCMOS camera delivers low noise, wide dynamic range, and high resolution for sensitive and quantitative measurements at short exposure time.
Application currently used: Fixed-cell assays, Live-cell assays, Complex cellular models, FRET assays, Phenotypic fingerprinting

Biotek MultiFlo™ FX
Manufacturer’s description: MultiFlo™ FX is an automated multi-mode reagent dispenser for 6- to 1536-well microplates. MultiFlo FX incorporates several unique technologies in its modular design, such as Parallel Dispense, RAD™ Random Access Dispense and the new, patent-pending AMX™ Automated Media Exchange modules to facilitate a variety of liquid handling applications from 2D and 3D cell culture to concentration normalization assays, ELISA, bead-based assays and more. A fully configured MultiFlo FX replaces up to five liquid handlers, saving space, time and instrumentation budgets.
Application currently used: Cell culture for automated reagent dispensing and washing (6to 384 wellplates)

TECAN HP D300 digital Dispenser
Manufacturer’s description: Fast and accurate determination of a candidate compound’s IC50 provides drug discovery biologists with valuable information for the development of new pharmaceuticals, yet traditional techniques are both time consuming and laborious, with no standardization across the industry. The HP D300 Digital Dispenser offers a simple method for streamlining your workflow, offering picoliter to microliter non-contact dispensing of small molecules in DMSO directly into your assay plate. Using HP’s Direct Digital Dispensing technology, this convenient benchtop solution requires almost no set up time, and single use T8 Dispenseheads virtually eliminate the risk of crosscontamination. It allows rapid delivery of any dose to any well, saving time, minimizing waste of valuable compounds and accelerating drug discovery.
Application currently used: Cell culture for delivery of pharmacological compounds in 96 and 384 well plates
Functional Genomics of Renal Cancer
Renal Cancer – 10 years of FunGeST expertise at the service of clinicians
Group Leaders
Actual Projects
J Zucman-Rossi (PUPH), B Beuselinck (PUPH Louvain)
Fundings: Inca
We aim to translate the molecular classification of clear cell renal cancer into genome based clinical trial int cooperation with Sautès-Fridman team at CRC and clinicans at HEGP and Louvain. We also search for biomarkers predicting response to anti-angiogenic thearapies.
Past Works
Recently, we described a molecular classification of clear cell renal cancer (ccRCCs) into four robust subgroups with distinctive characteristics on multi-omics analyses. These subgroups are defined by various levels of myc activation, angiogenic activation and immune response associated with outcome of metastatic ccRCC treated with sunitinib. This classification has future application in patient care, in particular to predict drug response.
Beseulinck et al, Clin Cancer Research 2015.
Publications
Fibroblast Growth Factor Receptor-2 Polymorphism rs2981582 is Correlated With Progression-free Survival and Overall Survival in Patients With Metastatic Clear-cell Renal Cell Carcinoma Treated With Sunitinib. Vanmechelen M, Lambrechts D, Van Brussel T, Verbiest A, Couchy G, Schöffski P, Dumez H, Debruyne PR, Lerut E, Machiels JP, Richard V, Albersen M, Verschaeve V, Oudard S, Méjean A, Wolter P, Zucman-Rossi J, Beuselinck B. Clin Genitourin Cancer. 2019 Apr;17(2):e235-e246. doi: 10.1016/j.clgc.2018.11.002. Epub 2018 Nov 16.
Polymorphisms in the Von Hippel-Lindau Gene Are Associated With Overall Survival in Metastatic Clear-Cell Renal-Cell Carcinoma Patients Treated With VEGFR Tyrosine Kinase Inhibitors. Verbiest A, Lambrechts D, Van Brussel T, Couchy G, Wozniak A, Méjean A, Lerut E, Oudard S, Verkarre V, Job S, de Reynies A, Machiels JP, Patard JJ, Zucman-Rossi J, Beuselinck B. Clin Genitourin Cancer. 2018 Aug;16(4):266-273. doi: 10.1016/j.clgc.2018.01.013.
Molecular Subtypes of Clear-cell Renal Cell Carcinoma are Prognostic for Outcome After Complete Metastasectomy. Verbiest A, Couchy G, Job S, Caruana L, Lerut E, Oyen R, de Reyniès A, Tosco L, Joniau S, Van Poppel H, Van Raemdonck D, Van Den Eynde K, Wozniak A, Zucman-Rossi J, Beuselinck B. Eur Urol. 2018 Oct;74(4):474-480. doi: 10.1016/j.eururo.2018.01.042. Epub 2018 Feb 17.
Pro-angiogenic gene expression is associated with better outcome on sunitinib in metastatic clear-cell renal cell carcinoma. Beuselinck B, Verbiest A, Couchy G, Job S, de Reynies A, Meiller C, Albersen M, Verkarre V, Lerut E, Méjean A, Patard JJ, Laguerre B, Rioux-Leclercq N, Schöffski P, Oudard S, Zucman-Rossi J. Acta Oncol. 2018 Apr;57(4):498-508. doi: 10.1080/0284186X.2017.1388927. Epub 2017 Nov 2.
Validation of VEGFR1 rs9582036 as predictive biomarker in metastatic clear-cell renal cell carcinoma patients treated with sunitinib. Beuselinck B, Jean-Baptiste J, Schöffski P, Couchy G, Meiller C, Rolland F, Allory Y, Joniau S, Verkarre V, Elaidi R, Lerut E, Roskams T, Patard JJ, Oudard S, Méjean A, Lambrechts D, Zucman-Rossi J. BJU Int. 2016 Dec;118(6):890-901. doi: 10.1111/bju.13585. Epub 2016 Aug 12.
RANK/OPG ratio of expression in primary clear-cell renal cell carcinoma is associated with bone metastasis and prognosis in patients treated with anti-VEGFR-TKIs. Beuselinck B, Jean-Baptiste J, Couchy G, Job S, De Reynies A, Wolter P, Théodore C, Gravis G, Rousseau B, Albiges L, Joniau S, Verkarre V, Lerut E, Patard JJ, Schöffski P, Méjean A, Elaidi R, Oudard S, Zucman-Rossi J. Br J Cancer. 2015 Nov 3;113(9):1313-22. doi: 10.1038/bjc.2015.352. Epub 2015 Oct 13.
Kidney cancer: Single nucleotide polymorphisms in mRCC-is their time up? Beuselinck B, Zucman-Rossi J. Nat Rev Urol. 2015 Aug;12(8):424-6. doi: 10.1038/nrurol.2015.149. Epub 2015 Jun 30. No
More publications
Our Lab
FunGeST Lab – A Multidisciplinary, Young & Motivated Team
Brief history of the team
FunGeST “Functional genomics of solid tumors”, directed by Jessica Zucman-Rossi, was created in 2005, as Inserm U674, it was renewed in 2009 and 2014 as UMR1162, ranked “incontournable” by Inserm as a mixed structure endorsed by four entities: Inserm, Universities Paris Diderot, Paris Descartes and Paris Nord. It is currently located at the University Hematology Institute site in a building managed by the CEPH (Centre d’Etude du Polymorphisme Humain, Foundation Jean Dausset). Since January 2019, the lab take part of Centre de Recherche des Cordeliers Research Center – Inserm UMR S1138 as team 28.
Jessica Zucman-Rossi, is professor of Medicine in Oncology (PUPHex) at university Paris Descartes and HEGP, full time for research. She directed the Inserm U674, U1162 and UMRS1138 – team 28 since 2007, was chairman of the Inserm scientific committee devoted to Oncology, Genetics and Bioinformatics from 2012 to 2016. She is editor at Journal of Hepatology (IF=12.4), executive secretary of the International Liver Cancer Association (ILCA). She published 157 original papers and has an international recognition in the field of genomics of human cancers and more specifically in liver tumors. Since January 2019 Pr Zucman-Rossi-Rossi is the director of the Cordeliers Research Center.
Our missions
Our mission is to develop basic genomic approaches based on human tumors analyses to identify new mechanisms of tumorigenesis and to transfer this knowledge into biomarkers and therapeutic targets that could be introduced in clinical care. In particular, we aim to identify new genomic alterations and mechanisms of carcinogenesis. We also aim to identify new risk factors and genetic predispositions promoting the development of tumors. Our general strategy is based on the omic’s analyses of large cohorts of patients with liver tumors (benign and malignant), mesothelioma and clear cell renal carcinoma. The group was pioneer in the elucidation of the molecular classification of benign and malignant liver tumors. Then we perform (1) functional validation using cell and animal models and (2) we translate our findings into the clinics to improve surveillance, diagnosis, prognosis, treatment and follow-up of the patients.
Our task forces
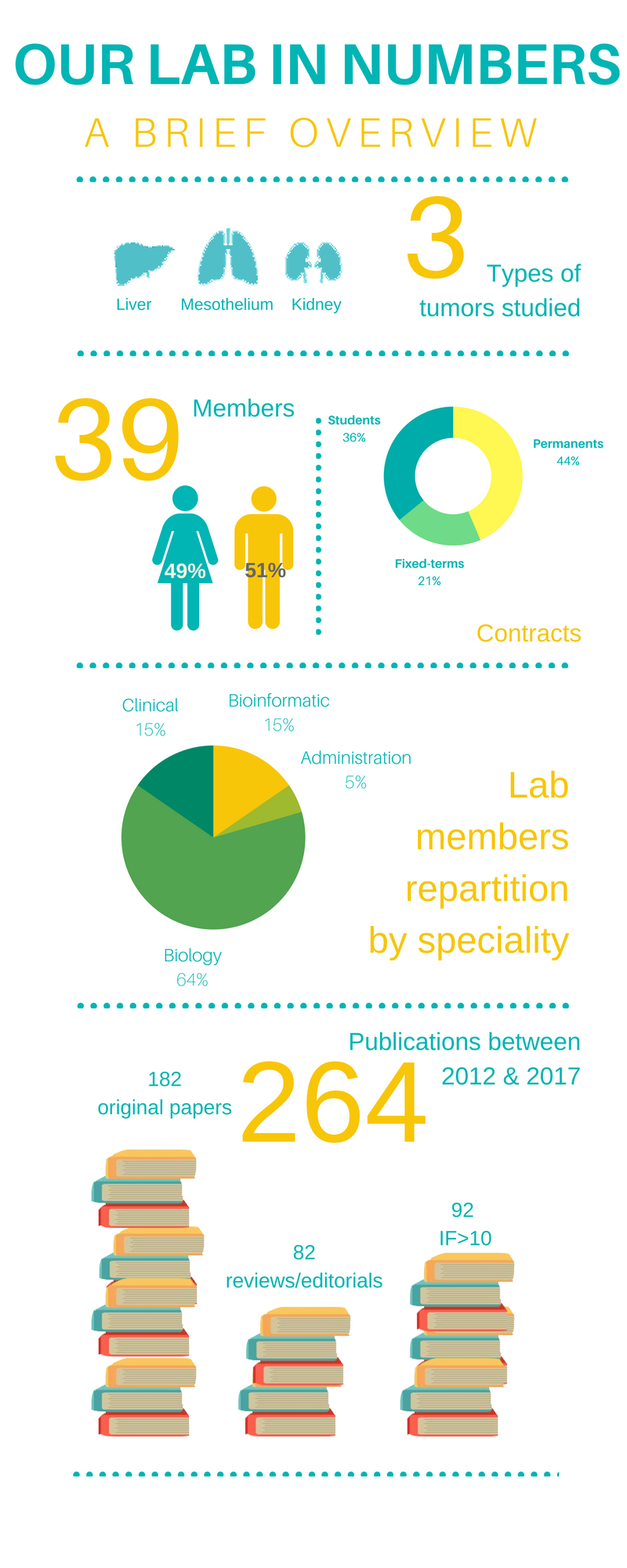
The team include a total of 39 peoples organized in 5 groups with their founded projects:
- “Functional Genomics of Liver tumors” led by Jessica Zucman-Rossi focusing in the characterization of genomic alterations in benign and malignant liver tumors, functional validation, identification of new risk factors and genetic predisposition and translation to the clinics.
- “Functional Genomics of Mesothelioma” led by Didier Jean (CR1 Inserm) focusing on the pathogenesis of mesothelioma, search for new therapeutic targets with genomic approaches.
- “Therapeutic Targets in liver tumors” led by Sandra Rebouissou (CR2 Inserm) focusing on the identification of therapeutic targets in liver tumors.
- “Computational Cancer Genome Analysis” led by Eric Letouzé (CR2 Inserm) focusing on the development and application of innovative strategies for integrative genomic data analysis.
- “Genomics of renal cancer” led by Jessica Zucman-Rossi & Benoit Beuselinck (Professor in Louvain)
Projects
FunGeST Projects
We aim to develop scientific projects with specific objectives to integrate innovative tumor genomic characterizations with metabolism and immune response to identify new biomarkers and therapeutic targets useful for the patients. To this aim, we focus on 3 major types of cancer: liver, mesothelioma and renal carcinoma, in close collaboration with clinicians and pathologists. Thanks to our future moving at the Centre de Recherche des Cordeliers our team will benefit from close collaborations with other teams involved in Onco-Immunology, Metabolism and developing innovative genomic approaches.